Parallel Programming Using R
Posted on August 6, 2018 by DEFTeamIn Parallel R Programming, the foreach package provides a new looping construct, which is used to execute a series of statements repeatedly. This looping construct performs the same work that
is done by other looping constructs such as for and while. But the main advantage of the foreach Parallel Processing in R is that it can execute the loop in parallel across multiple cores of a
system.
To execute a set of statements in Parallel R Programming using foreach package, we need
to
use doParallel R Programming package which is a parallel backend for foreach package. We need to
register
the doParallel to be used by foreach using registerDoParallel(cores=n) function. The argument to
that
function specifies how many cores that have to be used for performing the operation.
Comparison:
Time series data were taken in order to make a comparison between foreach Parallel R
Programming
package and normal R code(sequential). Time series data consists of 586 rows and 42 columns, where
each
row represents time series data for one location (time series object) and the column represents
month-wise
data. The chart below represents time taken by foreach package with different cores and without
using
foreach package i.e. sequential.
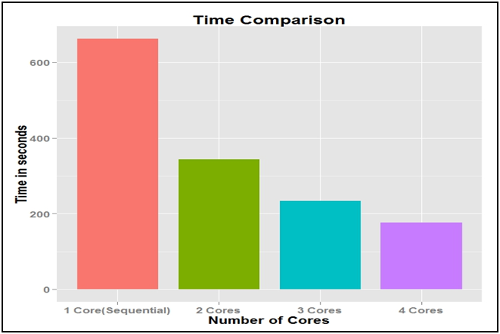
From the above result, we can see that using 4 cores in foreach Parallel R Programming
package is 3.77 times faster than R code which runs sequentially for time series data.
Here are the results when rolling forecast was done using foreach Parallel R
Programming
package.
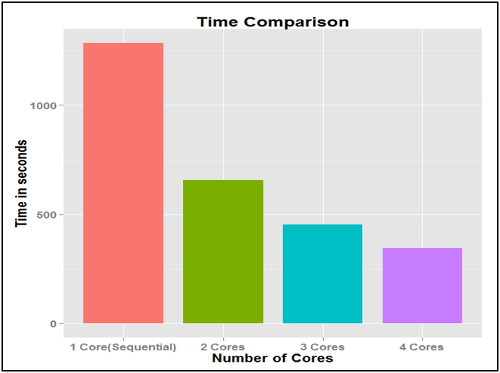
From the above result, we can see that using 4 cores in foreach Parallel R Programming
package is 3.73 times faster than R code which runs sequentially.
Above analysis was done on Desktop machine with 4 cores and 8GB of RAM.